#12,630
For the past 10 weeks we've been watching a growing, often severe, out-of-season H3N2 flu epidemic in Southeast China (see Macao, Hong Kong & Guangdong Province All Reporting Heavy Flu Activity) which appears to be hitting Hong Kong the hardest.
Yesterday, two Hong Kong University infectious disease experts suggested a recent mutation in the H3N2 virus may be one of the contributing factors to this summer flu epidemic (see Hong Kong: HKU Experts Call For Deploying Prophylactic Tamiflu To Avert Crisis).With much of the reportage in Chinese, we've had to deal with syntax challenged machine translations, but overnight the South China Morning Post reported:
Mutation may be behind surge in flu cases this summer, Hong Kong experts sayThe operative words in this headline being `may' and `could'.
Surge in number of cases could be due to variation in dominant strain, which may have rendered vaccines used in past two years ineffective
While several recent amino acid substitutions (including N121K) have turned up recently in H3N2 viruses in Hong Kong (and around the globe), it isn't at all clear how much of a factor they are in Hong Kong's current flu epidemic.
The H3N2 virus, which came to prominence as a pandemic strain in 1968 (supplanting 1957's pandemic H2N2), settled in the following year as a seasonal flu strain and remained the sole human influenza A strain until 1977, when H1N1 mysteriously reappeared after a 20 year absence.
Since then, H3N2 has co-circulated with (first) the old H1N1 strain (until 2009) and then the new H1N1pdm09 strain. Unlike in previous pandemics, the 2009 H1N1 virus did not supplant the existing (in this case, H3N2) influenza A virus.All of which makes H3N2 a bit long in the tooth, having been circulating in humans now for nearly 50 years. Over the past five decades it has had to reinvent itself innumerable times (via antigenic drift) to evade acquired immunity, resulting in an increasing number of subclades of the virus co-circulating around the globe.
Since 2009 seven distinct genetic groups have been defined for H3N2. While all belong to clade 3C, they are divided into three subdivisions; 3C.1 , 3C.2, and 3C.3, with last November's ECDC Influenza Virus Characterization Report stating:
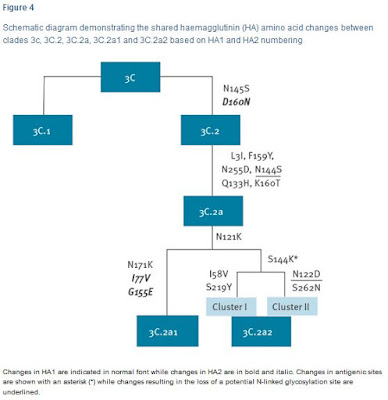
In 2014 three new subclades emerged, one in subdivision 3C.2, 3C.2a, and two in 3C.3, 3C.3a and 3C.3b , with subclade 3C.2a viruses dominating in recent months.Since then we've seen this newly dominant 3C.2a subclade further split into 3C.2a1 and 3C.2a2 (see Eurosurveillance: Emergence Of A Novel Subclade Of Seasonal A/H3N2 - London). The following Eurosurveillance chart shows the recent evolution of H3N2 clade 3C.
As you can see, both 3C.2a1 and 3C.2a2 share the N121K substitution, which is the `mutation' referenced in the SCMP article above. This amino acid substitution has increasingly been turning up in the literature over the past 6 months.
Last February, in a Eurosurveillance Rapid communication Interim estimates of 2016/17 vaccine effectiveness against influenza A(H3N2), Canada, January 2017 DM Skowronski et al. found:
Sequence analysis revealed substantial heterogeneity in emerging 3C.2a1 variants by province and over time. Adjusted VE was 42% (95% confidence interval: 18–59%) overall, with variation by province. Interim virological and VE findings reported here warrant further investigation to inform potential vaccine reformulation.In other words, they found the H3N2 vaccine effectiveness was a lackluster 42%, and that the H3N2 virus was evolving at a substantial rate. They write:
In sequencing analysis, we identified considerable diversity among circulating influenza A(H3N2) strains, including a mix of genetic variants that differed geographically and with time.
The majority (80%) of A(H3N2) viruses included in our VE analysis belonged to the newly emerging clade 3C.2a1, but with continuing genetic evolution compared with the vaccine strain. Almost all (95%) 3C.2a1 viruses had both the N171K and N121K mutations in site D that distinguish this clade.They end by saying:
Conclusion
Continued evolution in circulating 3C.2a variants and their derivatives, and the impact on vaccine protection, warrants ongoing monitoring to inform potential vaccine reformulation.
While a 40% VE is less than we'd like to see, it is far better than what we saw during the 2014-15 flu season when a late arriving `drifted' H3N2 virus practically negated that year's flu vaccine's effectiveness (see CDC HAN Advisory On `Drifted’ H3N2 Seasonal Flu Virus).
We've another study - referenced yesterday by the HKU experts - that found a far lower VE in Denmark for their H3N2 vaccine last winter, along with a very complex mixture of co-circulating H3N2 subclades.
J Clin Virol. 2017 Jun 29;94:1-7. doi: 10.1016/j.jcv.2017.06.007. [Epub ahead of print]
Changes in genetically drifted H3N2 influenza A viruses and vaccine effectiveness in adults 65 years and older during the 2016/17 season in Denmark.
Trebbien R1, Fischer TK2, Krause TG3, Nielsen L4, Nielsen XC5, Weinreich LS6, Lis-Tønder J7, Skov MN8, Christiansen CB9, Emborg HD3.
Abstract
RESULTS:
The genetic characterization revealed several genetically drifted viruses, which could be divided into four main clusters by the defining amino acid substitutions: 3C.2a/N121K/S144K, 3C.2a/T131K/R142K, 3C.2a1, and 3C.2a1/N121K.
Some of the drifted viruses appeared to be more prominent in vaccinated or non-vaccinated individuals, respectively. Overall the adjusted VE was 7.4% (95% confidence interval (CI): -6.0-19.2) among inpatients and 19.3% (95% CI: -5.7-38.4) among outpatients, respectively.
VE for the four main virus clusters was; cluster 3C.2a1: 38.8% (95% CI: -29.8-71.1), cluster 3C.2a/N121K/S144K: 9.2% (95% CI: -63.0-49.4), cluster 3C.2a/T131K/R142K: 19.0% (95% CI: -85.3-64.6), and cluster 3C.2a1/N121K: -12.2% (95%CI: -129.7-45.2).
CONCLUSIONS:
Several genetically drifted H3N2 viruses have been circulating in Denmark in the 2016-17 influenza season. An overall low VE was estimated and VE for the four main virus cluster indicate different VEs between the circulating drifted H3N2 viruses.
Copyright © 2017 Elsevier B.V. All rights reserved.
The VE against 3C.2a1 was nearly 40%, but for the remaining three drifted viruses was far lower, with the VE against the 3C.2a1/N121K virus almost nonexistent. These results all come from a cohort of over-65 aged patients, which tend to be the least protected by flu vaccines.
An equally dismal H3N2 VE was reported out of South Korea last winter (see PLoS One Interim estimates of the effectiveness of the influenza vaccine against A(H3N2) influenza in adults in South Korea, 2016–2017 season) although they did not provide any genetic breakdown of H3N2 subclades.Here in the United States, we saw a modest level of protection (34%) against H3N2, according to the CDC's MMWR report of June 30th.
This fall's northern hemisphere flu vaccine will use the same (A/Hong Kong/4801/2014-like) H3N2 virus as last year's formulation, but will add a new H1N1 strain. Its effectiveness in preventing H3N2 infection this year may well depend upon which `flavor' of the H3N2 virus visits your region this winter.2016–17 Influenza Vaccine Effectiveness
Data collected through the U.S. Influenza Vaccine Effectiveness Network during November 28, 2016–April 14, 2017, indicate that influenza vaccination this season reduced the overall risk for influenza-associated medical visits by 42% (95% CI = 35%–48%). Vaccine effectiveness against the predominant influenza A(H3N2) viruses was 34% (95% CI = 24%–42%) and vaccine effectiveness against influenza B viruses was 56% (95% CI = 47%–64%).
The evolution of flu viruses - particularly H3N2 - is so rapid that it is impossible to know with any certainty what subtype - much less what subclade - will be dominant six months from now.And the strain that is king of the viral mountain in North America in January may not be on top in Europe, or Asia. Which means, by the time flu arrives in your area, this fall's H3N2 vaccine component could do better, worse, or about the same as last year. Guaranteed.
I'll certainly get the flu vaccine this year (as I do every year), but given the increasing genetic diversity of the H3N2 virus and the severity of Hong Kong's outbreak, the advice offered by DM Skowronski et al. in the conclusion of their Eurosurveillance report above is something we probably all ought to take to heart this fall.
Given that a substantial proportion of vaccinated people may remain unprotected against influenza A(H3N2) illness, other adjunct measures should be considered to minimise associated morbidity and mortality, particularly among high-risk individuals.While it may not be as effective as we'd like, the flu vaccine – along with practicing good flu hygiene (washing hands, covering coughs, & staying home if sick) – remains your best strategy for avoiding the flu and staying healthy this winter.
And if you do get sick, early administration of antivirals - particularly for high-risk individuals - can make a real difference in severity, duration, and outcomes.